[CP-26] eScience, the Evolution of Science
Science - and research more broadly - face many challenges as its practitioners struggle to accommodate new challenges around reproducibility and openness. The current practice of science limits access to knowledge, information and infrastructure, which in turn leads to inefficiencies, frustrations and a lack of rigor. Many useful research outcomes are never used because they are too difficult to find, or to access, or to understand. New computational methods and infrastructure provide opportunities to reconceptualize how science is conducted, how it is shared, how it is evaluated and how it is reused. And new data sources changed what can be known, and how well, and how frequently. This article describes some of the major themes of eScience/eResearch aimed at improving the process of doing science.
Introduction
Topic Description:
- By Way of Introduction
- Major Themes of eScience
- Scaling Up Science
- Preserving Research Artifacts
- Packaging and Sharing Research Outcomes
- Workflows and Virtual Laboratories
- Describing Research Better
- "Live" Journals
- Conclusion
The very first academic journal was published in the year 1665: the Philosophical Transactions of the Royal Society, London. Coincidently, that was the same year as the great plague of London. New journals were added over time and the number of titles now is experiencing exponential growth, leading to a current total in excess of 25,000 peer reviewed journals (and another 50,000 scholarly periodicals) (Boon 2017). The number of published articles has shown a similar trend, achieving exponential growth over the last 25 years and around 2015 some lucky academic wrote the fifty millionth research article. The production rate of research is outstanding, but the mechanism for disseminating our knowledge is 350 years old and, to be frank, it shows. As a result, the reuse rate of all this hard-won knowledge is very poor. Academic papers are being published at the rate of 1.3 per minute and rising. In the social sciences, most are never cited (Van Noorden, 2017). Many great ideas, along with their supporting data and methods, go undiscovered because it is simply too hard to find them. And even when they are found, it is often too difficult to extract them from the dense text of the research paper. To give but one example, researchers in chemistry have found it necessary to develop an app that can recover data from the graphs embedded in published pdfs of journal articles! Reusing precious research outcomes developed by others in the conduct of their research remains a largely unsolved challenge.
A lot has changed since 1665: I no longer travel to work on a horse, nor get leeched when I have a fever. So why do we still communicate research via this outdated, static medium of the publication? The three pillars of science are: communicability, repeatability and refutability. Science cannot easily be repeated or refuted from a text in a library, though perhaps aspects of it can sometimes be communicated. One out of three is simply not good enough.
Computer science has tried to support the enterprise of research, developing tools such as databases, analysis methods, email, wikis, ontologies, data warehouses, containers and workflows, along with ever-faster processing and ever-larger storage. However, these tools are distinct, and poorly connected. Certainly, they have helped to make researchers more productive, but from a research communication perspective, they often make things worse, not better: they fragment the conduct of research across a dozen separate applications or more.
But imagine, if you will, a better container for a research artifact than paper (or pdf). Imagine that you could explore the actual methods, workflows and data used; where you could download self-describing data, and re-run analyses for yourself to validate them; where the use of research artifacts could be tracked automatically within communities, and artifacts could be discovered and accessed via a linked web of connections among all of the above. Imagine a published experiment where you could click on a graph to download the data, click on an equation to examine the code, click on the code and re-run the analysis. Imagine too, that all these connections worked both ways, so you could find all the methods that have been used on a dataset, or all the researchers who have validated an analysis. This is the vision that eScience aims to bring to life.
Currently, we might count ourselves lucky if we can find relevant papers by using keywords, but if you have you tried finding relevant datasets or analytical methods in the same manner, you will know how frustrating it is. Our searching tends to be restricted by our familiarity with certain domains, by knowing the right keywords to use, or by our social networks, or even by the journals that our university library subscribes to: all these restrictions constrain our ability to find and reuse the research of others.
The bottom line is this: for those millions of articles to be helpful to us, they need to be re-factored: that is, their contents prized apart and reorganized to maximize their reusability. This requires new ways of describing our research artifacts, with an emphasis on enabling discovery and re-use by other researchers–even researchers outside the originating community, and for tasks unforeseen by their creators.
Connected, scalable, live and reusable research is the overarching goal of eScience. Jim Grey (often regarded as the originator of eScience) famously said that: "everything about science is changing because of the impact of information technology." eScience began with the challenge of scaling up science applications using grid computing, but has evolved over time to become synonymous with any and all efforts to apply new information, and information technology to improve the science process, and outcomes. See Hey et al. (2009) for more on this new approach to science.
Some major themes of eScience are:
- Open science: science accessible to all with no barriers to uptake.
- Scaling up science: access to appropriately-sized computational infrastructure for all researchers.
- Making research artifacts persistent: identifiers and versioning to help us keep track of the things we make and use.
- Packaging and share research outcomes: containerization and libraries of pre-made software images for specific tasks,
- Workflows and virtual laboratories to capture and share entire experiments
- Describing research better: semantics of data and methods
- ‘Live’ research infrastructure that embeds data, code and workflow directly into the research article
A brief introduction to each of these themes in presented in turn below.
2.1 Open Science
The purpose of those first academic journals was to enable researchers to learn from the work of others without having to be physically present when findings were reported or scientific experiments conducted. The business models of publishing for profit or counting citations for promotion came much later. Open science aims to reduce the cost of participation in research, by making outcomes freely available to all, and at the same time to increase the transparency of research, by sharing code and data.
Technology now allows us now to create much more effective containers for all this precious research than paper, so let’s first explore what aspects of science (or research—the same ideas can apply outside of science) can be effectively ‘opened’. The open science movement has identified many components to the research enterprise that require a specific strategy to make them truly open, see the FOSTER portal for some useful discussion, and an excerpt is shown below in Table 1. A more detailed summary is provided by Knoth and Pontika (2015).
|
Aspiration |
Example |
|---|---|
|
Open-source Code |
Using platforms such as GitHub and Bitbucket as an open code repository as well as for version control. |
|
Open Data |
Using community-focused data infrastructure, such as the Neon Data Portal |
|
Open Review |
Journals that open share the details of both reviews and reviewers, and invite open discussion, such as the Semantic Web Journal |
|
Open Policies |
Clarity over funding decisions and priorities, such as OpenAire |
|
Open Tools, Workflows, and Virtual Laboratories |
Software environments that go beyond sharing the digital artifacts by also sharing how they all connect together into a repeatable sequence |
Within the GIScience community the Open Source Geospatial Foundation, colloquially known as OSGeo, has emerged in the last few years as a community dedicated to building open-source GISystems, such as QGIS and OSGeo4W. True to type, these codebases are fully open, as is the community who sustain them. It is therefore possible to use, examine and extend the code as you wish and share it on with whomever may want it. Some GIScience journals will now also publish code to accompany journal articles, which is a small but helpful step in the right direction.
2.2 Scaling up Science
Much of the early research in eScience concentrated on novel ways to make computing power accessible to all researchers (e.g. Foster and Kesselman 1999), not just those who could gain access to national High Performance Computing (HPC) facilities. This led to the creation of compute grids initially, and more latterly research clouds (both public and private). Typically, these platforms are built from commodity servers or even desktop machines, assembled into a single, large and virtual computer. Although they are not suitable for the highly coupled problems that we see in climate modelling or fluid dynamics, they are very useful in many genomics and spatial analysis applications, and far more performant than an office desktop. Most importantly, they provide a consistent computing platform for research communities to deploy consistent, shared software and data (see Sections 4 & 5). The task of migrating GIScience algorithms to the bigger HPC platforms remains an ongoing challenge (Shook et al., 2016).
2.3 Persistent Identifiers and Versioning
One of the key challenges in making ‘connected’ research is creating persistent digital identifiers that allow research artifacts to be uniquely identified and versioned. Establishing identifiers that persist and have meaning outside of a single institution or field is an organizational challenge. The current solution involves creating trusted authorities that can ‘mint’ new identifiers as a service, when required, and also validate what resource an identifier refers to. For example, ORCiD is a persistent identifier for researchers that subsumes any local identities a person may have (for example from past and present institutional affiliations). It is thus a more useful way of referring to an individual (Figure 1). ORCiD Identifiers remember who you are, even if you change your name, affiliation, even country.

Figure 1. A convenient reminder to use your ORCiD identifier. ORCiDs are an example of a persistent identifier that allows research activity to be connected to individuals regardless of their own professional mobility. Image source: author.
Persistent identifiers are also needed for the full range of digital research artifacts, including datasets, methods (code), instruments and even research activities. For example, consider Research Activity Identifiers, or RAID(link is external). Identifers like these are important because the URLs (uniform Resource Locators) used by the World Wide Web have turned out to be unreliable as a means to permanently identify the location of a resource: because resources move and network topologies change. To make things discoverable and reusable, we first need them to persist, and in an immutable way. See Klump and Huber (2017) for a useful summary of this topic.
2.4 Containerization
A further step towards repeatability is offered by "containerised" infrastructure such as Docker(link is external). The software container is a place to store a runtime image—a software bundle that includes application programs and often data as well. This image is created by serializing a working application—that is, writing out the memory holding the working software to storage. Then it can be easily shared. Upon receipt, it can be opened, "re-imaged" and run immediately. It will behave exactly the same as the original software did, thus it provides a very convenient way to "wrap-up" and share an experiment or piece of research with new users. Containers help support both repeatability and replicability and are mature enough now to be used reliably as part of a peer review process in science.
Scientific notebooks, such as the Jupyter Notebooks are popular variants of containerised infrastructure used in class and lab teaching throughout the world. They have several advantages: they are small and easy to deploy; they usually do not need much by way of computing resources to run; they combine code, data and description/documentation in a single environment; they help the user avoid overcomes installation and software integration issues; and they can be easily deployed at scale in the Cloud, say for teaching purposes. Also, they are typically small and self-contained, making them easier and quicker to deploy.
Large and diverse libraries of containerised applications are becoming available in several fields, including GIScience, allowing researchers to download and reuse the research tools of others. For example, this DockerImages list(link is external) represents a fast-growing library of open-source GIS functionality from the OSGeo community that can be easily reused.
2.5 Workflows and Virtual Laboratories
Workflow can be seen as an extension of containerization; as well as enabling the sharing of entire experiments, they go further and formally describe how the various software components are connected together. Workflow environments support the chaining together of analytical methods in a manner that completely describes all the steps in an experiment. It is in essence a language for specifying a directed graph, where the nodes are computational methods and the edges are links by which data (or control information) are passed in and out. See Figure 2 for an example. Often, this graph can be created via a visual interface, removing the need for programming skills. The graph can then be serialised—written out as a resource so that the analysis can be repeated, shared, and repurposed (Perkel, 2019).
The Galaxy workflow environment is perhaps the most well-known workflow engine and is used heavily in bio-informatics research. GeoVISTA Studio (Takatsuka and Gahegan, 2002) is an early example of a workflow and visual programming environment for geographical analysis and visualisation.
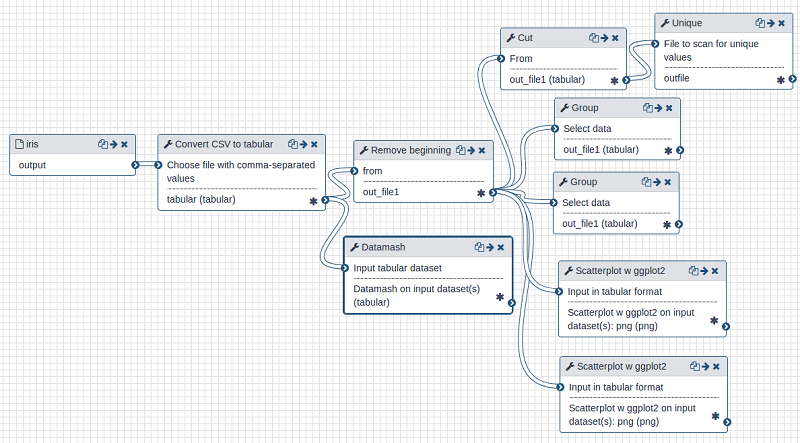
Figure 2. An executable workflow created in the Galaxy environment to input, combine and select data, that is then graphed as scatterplots. See Fouilloux et al, (2020) for more details. Source: author.
A natural extension of the ideas from cloud computing, containerization and workflows is the Virtual Laboratory, that provides a completely self-contained, environment combining application hosting, software modules and workflows and connections to relevant data collections. An excellent example is the Biodiversity and Climate Change Virtual Laboratory that supports some very sophisticated geospatial modelling and visualisation, but in a controlled environment that essentially wraps together all of the tools, data, methods and scripts used in analysis, serving as both shared resources and shared infrastructure to a research community.
2.6 Rich Descriptions of Data and Methods
The mode of communication in traditional journals is of course human to human. As journals have moved online, semantics have been increasingly used to describe the ‘container’ for the research article in progressively richer ways, via meta-data.
Research artifacts that can be ‘published’ or shared in some way need strong metadata to ensure they can be found using a search engine. Specific metadata related to publishing is often used to describe research articles, for example, and of course we have come to expect that such metadata is also used to power sophisticated search interfaces that help us to find useful content. Many publishing platforms for academic journals now also routinely use library metadata to enrich the description of each published article. For an example, see this useful summary from Crossref(link is external).
Many journals and journal indexing services now of course also use keywords to describe a research article, and some of these may be sourced from a controlled vocabulary or ontology. For example, GeoRef has been developing and maintaining structured vocabularies for geoscience publications since 1966. This kind of metadata starts to move beyond describing the article in general terms and begins to describe the contents themselves. The same goes for data and code, all of which can be richly described in terms of who created them, under what terms they can be reused and even what research questions they can help elucidate. See Davenhill (2011) for a more detailed and aspirational account of representing all science outcomes with metadata and Beard et al. (2010) for an early account using rich descriptive ontologies, workflows and shared code to empower an entire community. The Neon Data Portal(link is external) is a good example of how rich metadata can enable easier discovery of potentially useful data.
2.7 "Live" Journals
Perhaps the holy grail of reproducibility is a journal article that is also an executable experiment—describing an analysis in words, formulae and code, that can be repeated by the reader e.g. Chumbe et al. (2015). An excellent and recent example is the Physiome journal(link is external) that encourages authors to submit entire analytical workflows that accompany their more traditional article publications. Physiome evaluates submissions “to determine their reproducibility, reusability, and discoverability”. At a minimum, accepted submissions are guaranteed to be in an executable state that reproduces the modelling predictions in an accompanying primary paper, and are archived for permanent access by the community.” The journal uses well-established method libraries, process and data ontologies, common workflow descriptions and packaged data to deliver on its ambitious claims. It is the culmination of many years of collaborative research within a segment of the bioengineering community. Could we not do the same, using the codebase of an open-source GIS?
If we take the challenges of eScience seriously, we will work to improve the way our academic outcomes are represented, communicated, archived, discovered, reused and valued by changing our own behavior. This includes redoubling our efforts to make our own research open to all, and as transparent as repeatable as we can manage. It also may involve challenging the status quo in academia, which does not always incentivise the most helpful and productive behaviours. At the very least, we need to work towards a system that rewards ‘good’ behaviour. One small step might be to afford published code and data the same status as a research article, with peer review and citation counts used as a measure of quality.
If we meet these challenges, we will make science—and indeed all research—more discoverable, more reproducible, more honest, and ultimately more useful. If we don’t, we will hand over a bigger mess to the next generation of researchers than the one we ourselves inherited.
External resources
- Beard, D.A., Britten, R., Cooling, M.T., Garny, A., Halstead, M.D., Hunter, P.J., Lawson, J., Lloyd, C.M., Marsh, J., Miller, A. and Nickerson, D.P. (2009). CellML metadata standards, associated tools and repositories. Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences, 367:1895, pp.1845-1867.
- Chumbe, S., Kelly, B., & MacLeod, R. (2015). Hybrid journals: Ensuring systematic and standard discoverability of the latest open access articles. The Serials Librarian: From the Printed Page to the Digital Age, 68(1-4), 143-155. DOI:10.1080/0361526X.2015.1016856
- Davenhall, C. (2011). The DCC Digital Curation Reference Manual, Instalment on Scientific Metadata. Online publication available from: http://www.dcc.ac.uk/sites/default/files/documents/Scientific%20Metadata_2011_Final.pdf
- Foster, I., and Kesselman, C. (1999). The Grid: Blueprint for a New Computing Infrastructure. Morgan Kaufmann Publishers. ISBN 978-1-55860-475-9.
- Fouilloux, A., Goué, N., Barnett, C., Maroni, M., Nahorna, O., Clements, D., Hiltemann, S. 2020. Galaxy 101 for everyone (Galaxy Training Materials). /training-material/topics/introduction/tutorials/galaxy-intro-101-everyone/tutorial.html. Online; accessed Fri. Sep. 11, 2020.
- Hey, T., Tansley, S. and Tolle, K. Eds. (2009). The Fourth Paradigm: Data-Intensive Scientific Discovery, Redmond, VA: Microsoft Research, 2009, ISBN 978-0-9825442-0-4, http://fourthparadigm.org
- Klump, J., and Huber, R. (2017). 20 Years of Persistent Identifiers – Which Systems are Here to Stay?. Data Science Journal, 16, 9. DOI: 10.5334/dsj-2017-009.
- Knoth, P. and Pontika, N. (2015) The Open Science taxonomy. www.fosteropenscience.eu/resources.
- Perkel, J. M. (2019). Workflow systems turn raw data into scientific knowledge. Nature, 573, 149-150. DOI: 10.1038/d41586-019-02619-z.
- Shook, E., Hodgson, M. E., Wang, S., Behzad, B., Soltani, K., Hiscox, A. and Ajayakumar, J. (2016). Parallel cartographic modeling: a methodology for parallelizing spatial data processing. International Journal of Geographical Information Science 30 (12):2355–2376. DOI: 10.1080/13658816.2016.1172714.
- Takatsuka, M. and Gahegan, M. (2002). GeoVISTA Studio: a codeless visual programming environment for geoscientific data analysis and visualization. Computers and Geosciences 28(10):1131-1144 2002. DOI: 10.1016/S0098-3004(02)00031-6
- Van Noorden, R. (2017). The science that’s never been cited. Nature, 552, 162-164. DOI: 10.1038/d41586-017-08404-0
Learning outcomes
-
96 - Explain, with examples, how all manner of research outcomes could be shared more effectively
Explain, with examples, how all manner of research outcomes could be shared more effectively
-
97 - Propose a set of minimum standards for the more effective sharing of GIS code and data
Propose a set of minimum standards for the more effective sharing of GIS code and data
-
98 - Contrast the approach to research afforded by eScience ideas with the approach you see practiced currently in GIScience and Geography
Contrast the approach to research afforded by eScience ideas with the approach you see practiced currently in GIScience and Geography
Outgoing relations
- [CP-26] eScience, the Evolution of Science is subconcept of Computing Platforms
Contributors
-
Gahegan, Mark. (2020). eScience: the Evolution of Science. The Geographic Information Science & Technology Body of Knowledge (3rd Quarter 2020 Edition), John P. Wilson (ed.).
This entry was first published on September 24, 2020. No earlier editions exist.
https://doi.org/10.22224/gistbok/2020.3.8